Infection with SARS-CoV-2 results in antibodies against common colds
Infection by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) provides immunity against seasonal human coronaviruses (HCoVs). Whether SARS-CoV-2 infection causes a boost in the preexisting HCoV-specific antibodies or elicits cross-reactive β-CoV antibodies that target conserved epitopes is unknown.
A study published in the journal Science Advances uses electron microscopy-based polyclonal epitope mapping (EMPEM) methodology to elucidate the spike epitopes of β-HCoV targeted by preexisting serum antibodies and compares them to the epitopes targeted by antibodies elicited by SARS-CoV-2 infection using electron microscopy-based polyclonal epitope mapping (EMPEM) methodology.
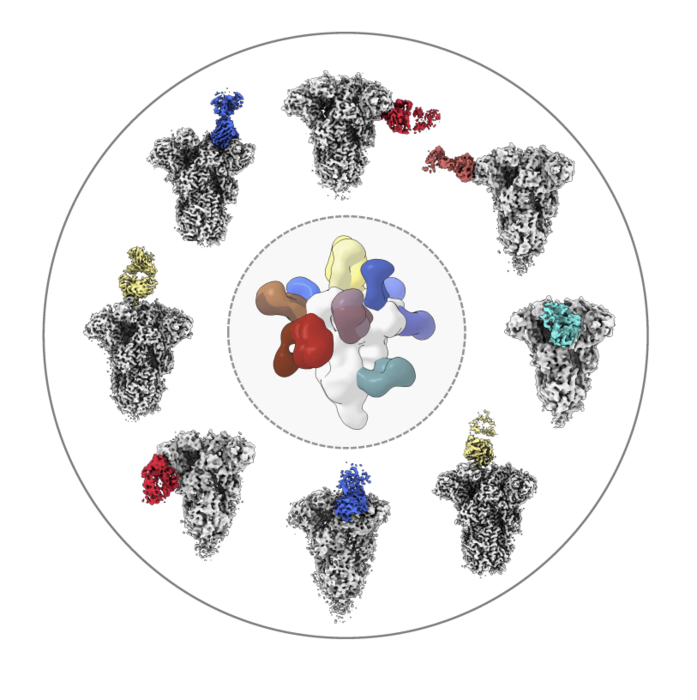
Researchers analyzed the structures of antibodies (colored) from healthy donors as the molecules bound to the spike protein from common coronaviruses like OC43 (shown here in white/gray). Image Scripps Research
Human coronaviruses
One-third of the common cold infections are caused by HCoVs. Two αHCoVs, 229E and NL63, and two βHCoVs, OC43 and HKU1, are endemic in the human population. A majority of the population acquires HCoV infections by the age of 15 years. However, the infection rate and prevalence differ between different geographical locations.
Since the population is exposed to HCoVs, most individuals have antibodies against these viruses. The antibodies target the spike protein and the nucleocapsid protein of the viruses. However, the antibody levels wane over time, and reinfections can happen, sometimes within a year.
The β-CoVs also include the highly pathogenic Middle East respiratory syndrome (MERS) CoV, SARS-CoV, and SARS-CoV-2. The coronavirus disease 2019 (COVID-19) pandemic is the result of the high pathogenicity and transmissibility of SARS-CoV-2.
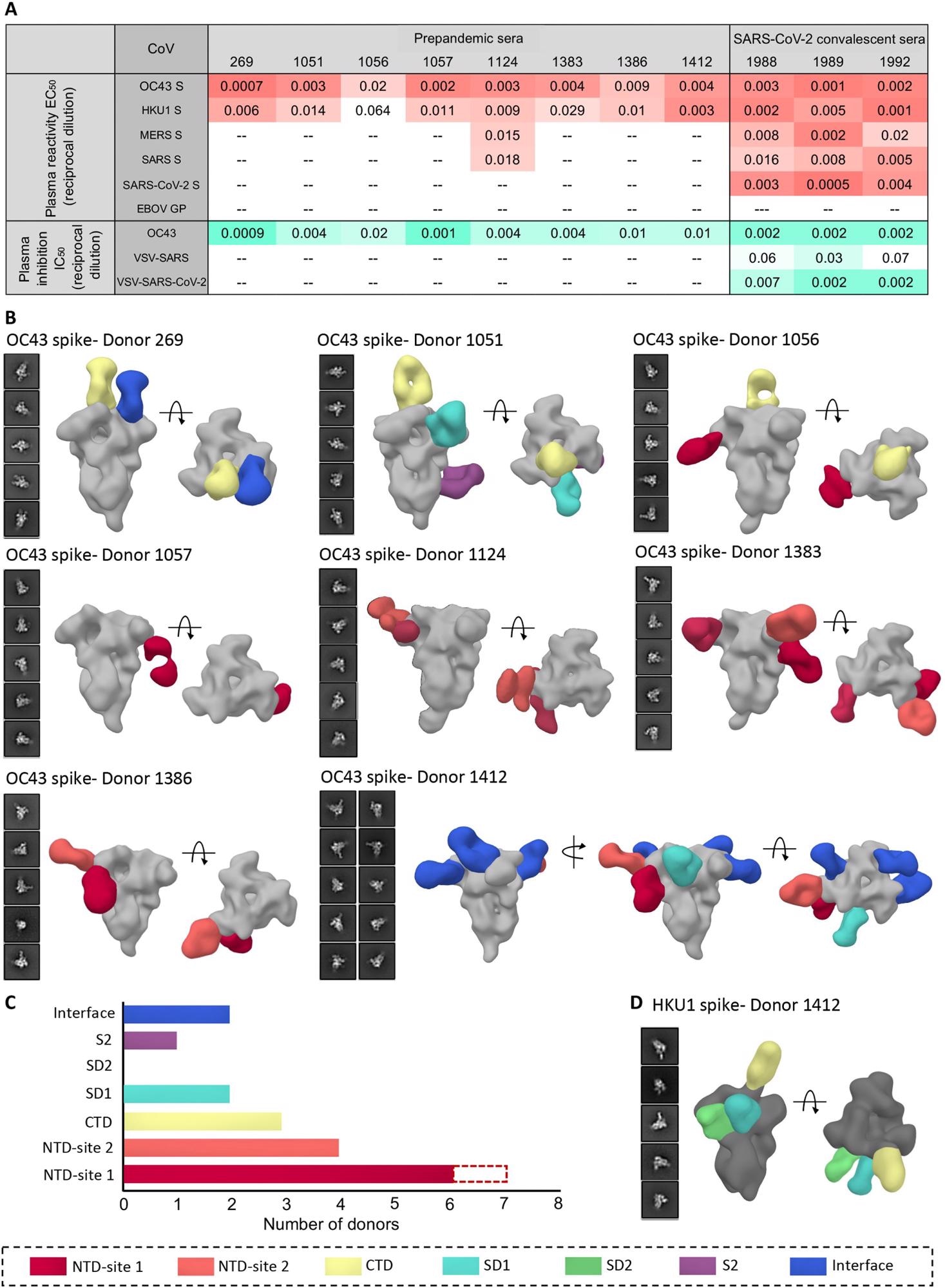 Human serum reactivity to β-CoV spikes. (A) ELISA EC50 binding titers to OC43, HKU1, MERS, SARS, and SARS-CoV-2 spikes and median inhibitory concentration (IC50) neutralization titers against OC43 virus and vesicular stomatitis virus (VSV)–pseudotyped SARS or SARS-CoV-2 virus for PP sera from eight healthy donors and SC sera from three SARS-CoV-2 donors. Ebola virus glycoprotein (EBOV GP) was used as a negative control for detecting nonspecific serum binding. Serum EC50 or IC50 titers are color-coded in gradients of orange or aquamarine, respectively. (B) Representative two-dimensional (2D) classes and side and top views of composite figures from ns-EMPEM analysis of polyclonal Fabs from eight PP sera with the OC43 spike. (C) Bar graph summary of OC43 spike epitopes targeted by PP donor sera. Antibodies to NTD-site 1 were observed in 2D class averages for donor 269 but did not reconstruct in 3D, as indicated by dotted lines. (D) Composite figures from ns-EMPEM analysis of polyclonal Fabs from donor 1412 with the HKU1 spike. The Fabs in (B) and (D) are color-coded on the basis of their epitope specificities as indicated at the bottom. OC43 or HKU1 spikes in (B) and (D) are represented in light gray or dark gray, respectively.
Human serum reactivity to β-CoV spikes. (A) ELISA EC50 binding titers to OC43, HKU1, MERS, SARS, and SARS-CoV-2 spikes and median inhibitory concentration (IC50) neutralization titers against OC43 virus and vesicular stomatitis virus (VSV)–pseudotyped SARS or SARS-CoV-2 virus for PP sera from eight healthy donors and SC sera from three SARS-CoV-2 donors. Ebola virus glycoprotein (EBOV GP) was used as a negative control for detecting nonspecific serum binding. Serum EC50 or IC50 titers are color-coded in gradients of orange or aquamarine, respectively. (B) Representative two-dimensional (2D) classes and side and top views of composite figures from ns-EMPEM analysis of polyclonal Fabs from eight PP sera with the OC43 spike. (C) Bar graph summary of OC43 spike epitopes targeted by PP donor sera. Antibodies to NTD-site 1 were observed in 2D class averages for donor 269 but did not reconstruct in 3D, as indicated by dotted lines. (D) Composite figures from ns-EMPEM analysis of polyclonal Fabs from donor 1412 with the HKU1 spike. The Fabs in (B) and (D) are color-coded on the basis of their epitope specificities as indicated at the bottom. OC43 or HKU1 spikes in (B) and (D) are represented in light gray or dark gray, respectively.
Spike protein
The spike protein binds to the angiotensin-converting enzyme 2 (ACE2) receptor of the host cells. This binding mediates cellular entry. Therefore, the spike protein decides the host range and types of cells infected (cell tropism). It is also the key target for neutralizing antibodies and consequently the major antigen for vaccine development.
The SARS-CoV-2 spike is 69.2% similar to the SARS spike and 27.2% similar to the OC43 spike. It shares little similarity with the spike of other β-CoVs. Even so, preexisting immunity against HCoVs correlates with COVID-19 disease outcomes. This could be due to a boosting of anti-HCoV-spike-specific antibodies after SARS-CoV-2 infection. Alternatively, this could be because SARS-CoV-2 infection elicits antibodies that cross-react with the HCoV spike.
Interestingly, individuals infected with SARS-CoV-2 having high anti-SARS-CoV-2 antibody levels also have increased levels of anti-β-HCoV antibodies.
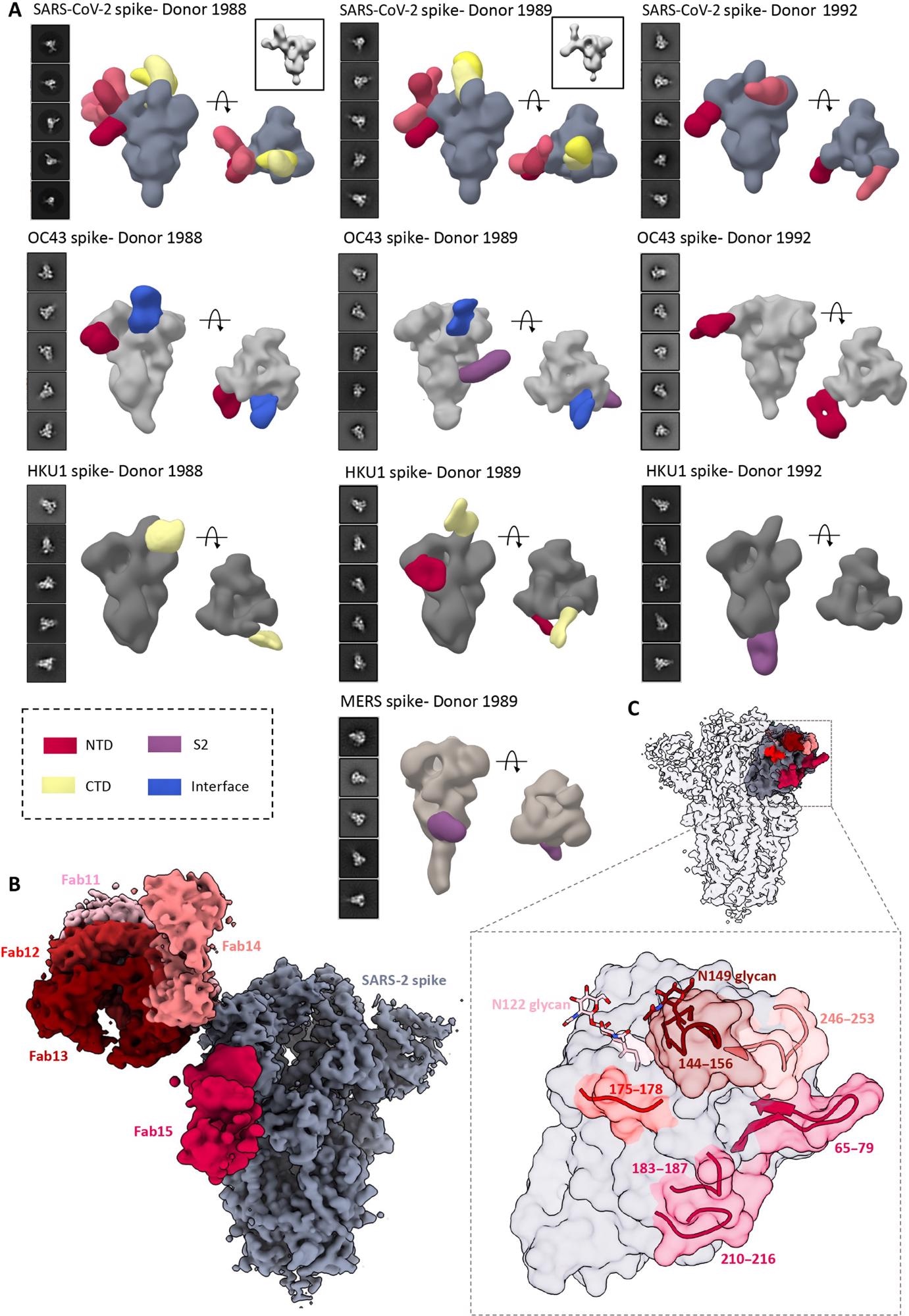
4. ns- and cryo-EMPEM analysis of polyclonal Fabs from SC donor sera. (A) Representative 2D classes and side and top views of composite figures from ns-EMPEM analysis of polyclonal Fabs from three SC donors complexed with β-CoV spikes. The donor numbers along with the corresponding CoV spikes are indicated above each panel in (A). The Fabs are color-coded on the basis of their epitope specificities as indicated at the bottom left. SARS-CoV-2, OC43, HKU1, and MERS spikes are represented in slate gray, light gray, dark gray, and beige, respectively. Three-dimensional reconstructions displaying potential self-reactive antibodies are shown in gray on the top right corners for both donors 1988 and donor 1999 in complex with SARS-CoV-2 spike. (B) Composite figure showing five unique antibody classes, Fab11 to Fab15 colored in shades of red, to SARS-CoV-2 spike NTD reconstructed using cryo-EMPEM analysis of polyclonal Fabs from donors 1988 and 1989 complexed with SARS-CoV-2–stabilized spikes. (C) Surface representation of SARS-CoV-2 spike showing epitopes of Fabs 11 to 15 from (B) on a single NTD (slate gray) with a zoomed-in view displaying the loop residues comprising each epitope. Loop 144 to 156 with the N149 glycan forms an immunodominant element commonly targeted by Fabs 11 to 14. The sub-epitope colors correspond to each Fab shown in (B).
Sera analyses
The investigators of this study generated soluble protein domains of the spike proteins of HKU1, OC43, SARS, MERS, and SARS-CoV-2. These constructs were characterized using negative stain electron microscopy (ns-EM).
Blood samples were collected from eight donors before the COVID-19 pandemic and from three donors who experienced SARS-CoV-2 infection after the pandemic. The sera were tested for anti-spike antibodies using an enzyme-linked immunosorbent assay (ELISA).
All eight pre-pandemic (PP) sera had anti-OC43 spike antibodies. Anti-HKU1 spike antibodies were present at very low levels in all eight sera. This may be because OC43 is prevalent throughout the world, while HKU1 is less prevalent. None of these sera had anti-SARS-CoV-2 spike antibodies. One serum sample had low levels of anti-SARS and anti-MERS spike antibodies.
The three SARS-CoV-2 convalescent (SC) sera had high levels of anti-SARS-CoV-2 spike antibodies. They also had anti-SARS, anti-MERS, anti-OC43, and anti-HKU1 spike antibodies. Thus, SARS-CoV-2 infection can elicit some level of cross-reactive antibodies against the β-CoV spike proteins.
Next, the investigators determined if PP and SC sera neutralized OC43 virus and SARS and SARS-CoV-2 pseudoviruses. As expected, none of the PP sera neutralized the SARS or SARS-CoV-2 pseudovirus. The SC sera neutralized the OC43 virus and SARS and SARS-CoV-2 pseudoviruses.
Structural analyses
The investigators determined the specific epitopes targeted by the anti-spike antibodies in the PP sera by ns-EMPEM. The structures of antibody-spike complexes were analyzed. A portion of the antibodies was used for this analysis, not the entire antibody.
The investigators performed high-resolution cryo-EMPEM studies with OC43 spike using three PP sera.
The investigators also explored the nature of anti-spike antibodies following SARS-CoV-2 infection. The three SC sera were screened for anti-SARS-CoV-2 spike by ns-EMPEM.
Structural mapping of anti-SARS-CoV-2 spike residues that are similar or identical to at least three of the four other β-CoVs showed several conserved patches in the S2 subunit of the spike protein that can elicit cross-reactive antibodies. So, the S2 subunit is a target for cross-reactivity across β-CoVs.
The PP and SC sera had anti-OC43 spike antibodies. Two SC sera had antibodies against the N-terminal domain (NTD) site 1, two SC sera had antibodies against the interface, and one SC serum had antibodies against the S2 subunit. The anti-S2 antibody-targeted the helix 1014 to 1030, a portion of the spike protein highly conserved across the β-CoVs. PP sera with high levels of anti-OC43 antibodies also had anti-SARS-CoV-2 antibodies that did not correlate with protection against SARS-CoV-2 infection.
A SARS-CoV-2 infection resulted in an increase in the levels of anti-HKU1 spike antibodies. These antibodies were specific to the C-terminal domain (CTD) and/or the NTD and S2 domain.
Antibodies against the S1 subunit were observed in both PP and SC sera but those against the S2 subunit were enriched in SC sera.
Conclusion
A SARS-CoV-2 infection induces cross-reactive antibodies to conserved β-CoV spike epitopes at the same time back-boosting some HCoV anti-spike antibodies. This cross-boosting correlates with COVID-19 pathogenesis. This cross-boosting may also provide immunity to seasonal HCoVs to the individuals who are vaccinated or previously infected with SARS-CoV-2.
- Sandhya B, Aleksandar A, Nurgun K, et al. (2022). Structural mapping of antibody landscapes to human betacoronavirus spike proteins. Science Advances. 8(18):eabn2911. doi:10.1126/sciadv.abn2911, https://www.science.org/doi/10.1126/sciadv.abn2911
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Antigen, Assay, Blood, Cell, Cold, Common Cold, Coronavirus, Coronavirus Disease COVID-19, covid-19, Electron, Electron Microscopy, Enzyme, Glycan, Glycoprotein, Helix, immunity, Microscopy, Pandemic, Protein, Pseudovirus, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Stomatitis, Syndrome, Vaccine, Virus

Written by
Dr. Shital Sarah Ahaley
Dr. Shital Sarah Ahaley is a medical writer. She completed her Bachelor's and Master's degree in Microbiology at the University of Pune. She then completed her Ph.D. at the Indian Institute of Science, Bengaluru where she studied muscle development and muscle diseases. After her Ph.D., she worked at the Indian Institute of Science, Education, and Research, Pune as a post-doctoral fellow. She then acquired and executed an independent grant from the DBT-Wellcome Trust India Alliance as an Early Career Fellow. Her work focused on RNA binding proteins and Hedgehog signaling.
Source: Read Full Article


