New study of age-related macular degeneration finds causal genes, dispels previous assumptions
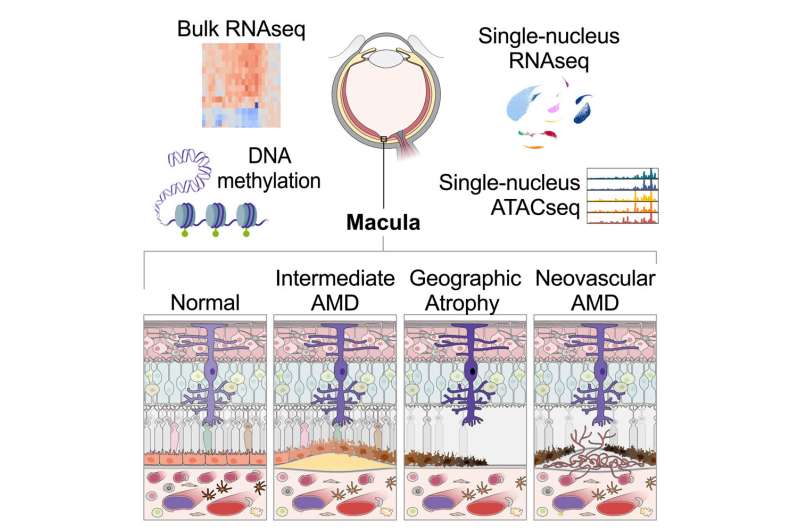
A study led by the research center of Genentech in South San Francisco, published in Cell Genomics, has looked for drug targets that could address age-related macular degeneration (AMD), a condition at the center of vision problems for 200 million people worldwide. AMD can result in blindness.
In the paper “A systems biology approach uncovers novel disease mechanisms in age-related macular degeneration,” the researchers describe the steps they took to identify genes targetable by treatment with a molecular atlas of AMD pathology development stages.
The study resulted in the identification of 23 significant genome-wide loci that are differentially methylated in AMD. Over 1,000 differentially expressed genes were found across disease stages and distinct Müller cell states in AMD-affected eyes. The research highlights causal gene upregulations and underlying genetic risks for AMD.
Researchers generated bulk-tissue and single-cell transcriptomics and epigenomics data from 85 unique human donor eyes to understand the molecular changes occurring as AMD develops. They analyzed tissue transcriptomes of early, intermediate, and two types of advanced-stage AMD.
One of the intriguing finds of the study is the different clusters of Müller glia cell states—basal, AMD, and gliotic. A total of 62% of the basal Müller cluster came from controls, and 80% of the AMD Müller cluster came from AMD donors. The study authors point out that although Müller gliosis is a common feature in retinal diseases and injury, the AMD Müller cluster did not show higher expression of gliosis markers (upregulated glial fibrillary acidic protein). This observation in actual human disease states differs from retinal injury models often used in AMD translational research.
The research also points out that the gliotic state appears to be a crucial intermediate between normal Müller glia and the stem cell identity in retinal regeneration. This suggests that as retinal regeneration research translates into therapeutic strategies, a deeper understanding of the disease state Müller glia is needed, as potential therapeutics designed to reprogram the basal or gliotic Müller may not be suitable for the AMD-like state.
Analysis between control and AMD donors found no genome-wide significant differences. There were no overt differences between control and AMD samples at cell type or subtype levels.
In contrast to previous research in 2018 by John Hopkins researchers, “ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration,” published in Nature Communications, in which the AMD disease state was found to be associated with a global reduction of open chromatin, the Genentech researchers did not detect a corresponding shift in chromatin accessibility.
Based on the previously reported observation, the current study expected to observe disease-related differences in chromatin accessibility with a higher resolution of detail. However, the current study did not see a replication of the previous research findings of a shift in chromatin accessibility either globally or at specific loci, suggesting changes in chromatin accessibility in the past bulk analysis may have been simply due to cell death in the sample with a confounding of nuclei from non-disease-related cell types.
The Genentech researchers, along with colleagues from the University of Utah and the State University of New York at Buffalo, have dispelled a few past assumptions and confounding research attempts and identified a robust number of relevant gene mechanisms related to AMD that will likely form the basis for future research on the subject.
More information:
Luz D. Orozco et al, A systems biology approach uncovers novel disease mechanisms in age-related macular degeneration, Cell Genomics (2023). DOI: 10.1016/j.xgen.2023.100302
Journal information:
Nature Communications
,
Cell Genomics
Source: Read Full Article


